
DYOGEN group
web-code version: 2017-06-13
database version: 02.01
 Contact us.
Contact us.

|

|

|
|
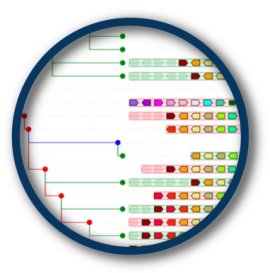
| Root species for KH2012:KH.L106.16 (Cirobu) | AlignView depth | PhyloView depth | |
|---|---|---|---|
| 34_Deuterostomia |  ~10 Mya ~10 Mya |
33 species  |
1442 homologs (oldest homol. - dupl. node)  1434 homologs (dupl. node)  983 homologs (dupl. node)  976 homologs (dupl. node)  969 homologs (dupl. node)  895 homologs (dupl. node)  673 homologs (dupl. node)  250 homologs (dupl. node)  181 homologs (dupl. node)  153 homologs (dupl. node)  68 homologs  |
| 30_Chordata |  ~10 Mya ~10 Mya |
29 species  |
42 homologs (dupl. node)  23 homologs  |
| 28 |  ~10 Mya ~10 Mya |
27 species  |
22 homologs  |
| 16_Ascidiacea |  ~10 Mya ~10 Mya |
17 species  |
21 homologs (dupl. node)  16 homologs  |
| 15_Phlebobranchia |  ~10 Mya ~10 Mya |
7 species  |
7 homologs  |
| 11_Ciona |  ~10 Mya ~10 Mya |
3 species  |
3 homologs  |
| Cirobu |  ~0 Mya ~0 Mya |
1 species  |
1 homolog  |
-evalue <Real>
Expectation value (E) threshold for saving hits
Default = '10'
-word_size <Integer, >=2>
Word size for wordfinder algorithm
Default = '3'
-gapopen <Integer>
Cost to open a gap
Default = '11'
-gapextend <Integer>
Cost to extend a gap
Default = '1'
-matrix <String>
Scoring matrix name (normally BLOSUM62)
-threshold <Real, >=0>
Minimum word score such that the word is added to the BLAST lookup table
Default = '11'
-window_size <Integer, >=0>
Multiple hits window size, use 0 to specify 1-hit algorithm
Default = '40'